ROTDIF
Matlab program aiming at the determination of the overall rotational diffusion tensor of a molecule from spin-relaxation data. Features isotropic, axially-symmetric, and fully anisotropic models for the overall motion. Reference: Fushman et al., Biochemistry 38 (1999) 10225-10230; Varadan et al., JMB 324 (2002) 637; Walker et al. J. Magn. Reson. (2004) 168, 336; Fushman et al, Progress NMR Spectroscopy, (2004) 44, 189.
This program is currently available from Prof. David Fushman’s group.
SPINRELAX
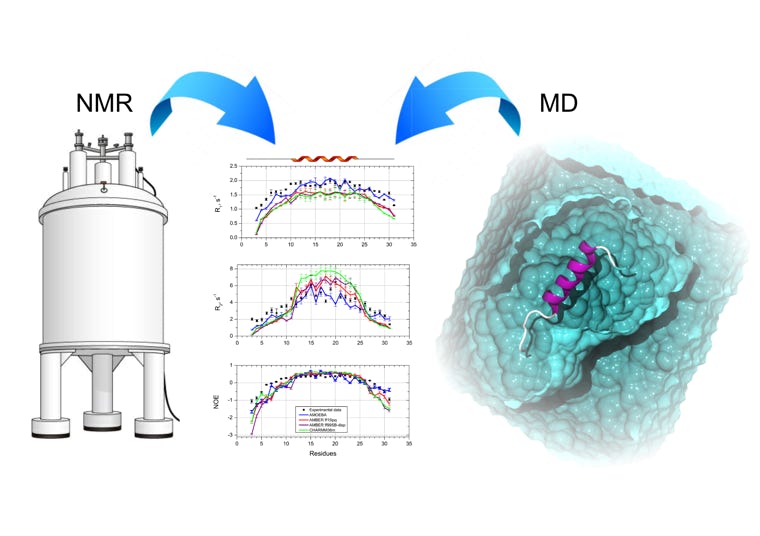
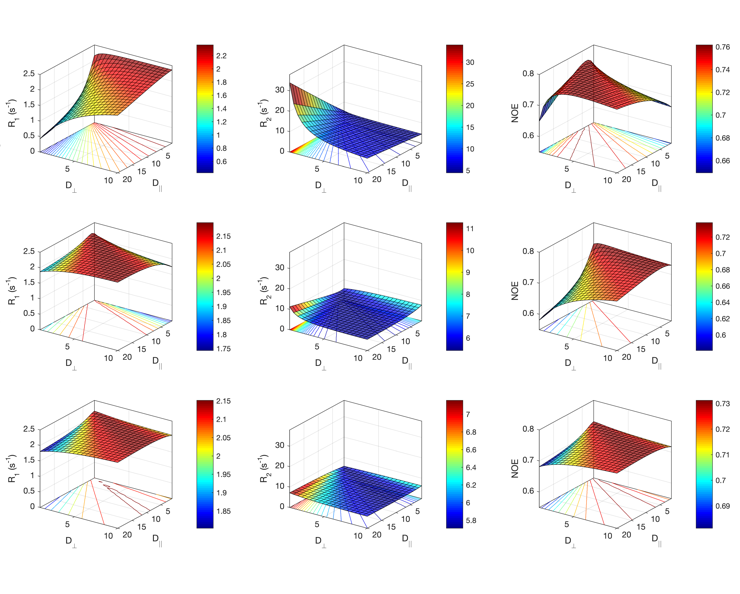
This program aims at the calculation of NMR spin relaxation rates R1, R2 and NOEs from one or several molecular dynamics trajectories. Different parameters may be tuned like temperature, isotropic diffusion, magnetic fields.
Additionally, experimental relaxation data may be fitted to derive an optimized CSA, isotropic rotational diffusion or S2. References: Ab Initio Prediction of NMR Spin Relaxation Parameters from Molecular Dynamics Simulations or Accurate Prediction of Protein NMR Spin Relaxation by Means of Polarizable Force Fields.
This program is freely available Po-Chia’s repository